Abstract
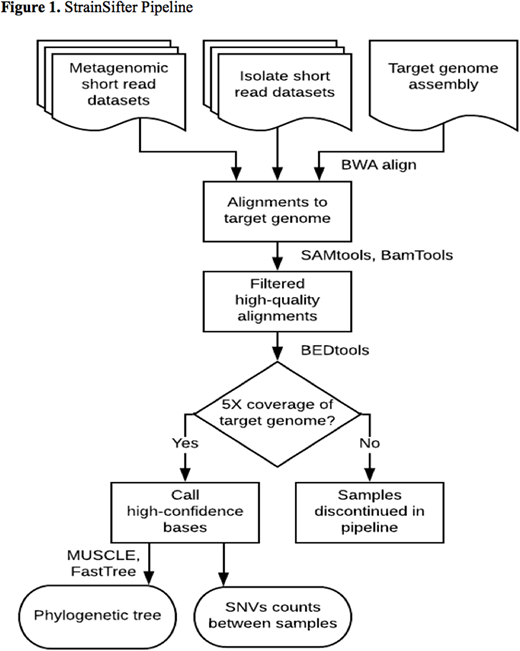
Recipients of hematopoietic stem cell transplantion (HCT) are at high risk for bloodstream infections (BSI); culture-based and marker gene sequencing approaches have been previously applied to interrogate the intestine as a reservoir of these pathogens. Identifying the source of bloodstream infections can impact infection control and prevention practices such as removing central venous catheters and placing patients in isolation. To evaluate the genomic concordance of bloodstream pathogens and bacterial strains within the intestinal microbiome, we developed StrainSifter (Figure 1), a streamlined bioinformatic pipeline using whole genome shotgun sequencing to compare nucleotide variation between bacterial isolate strains and metagenomes. We applied StrainSifter to bloodstream isolates and stool metagenome samples from a cohort of 30 HCT recipients with bloodstream infections at a single academic center. StrainSifter is designed to 1) identify single nucleotide variants (SNVs) between isolate and metagenomic short reads aligned to assembled draft genomes using stringent alignment, coverage, and variant frequency criteria, and 2) output both phylogenetic trees and SNV counts for clinical and epidemiologic strain typing and comparison. Using StrainSifter, we identified enteric BSI isolates - including Enterococcus faecium, Klebsiella pneumoniae, Escherichia coli, and Streptococcus mitis - that were identical or nearly identical with those in the gut microbiota. We also identified highly concordant strains of Staphylococcus epidermidis and Pseudomonas aeruginosa between the bloodstream and gut microbiome, an unexpected finding for typically non-enteric organisms. Although intestinal domination by pathogenic bacteria preceded BSI with E. faecium and E. coli as previous studies have demonstrated, we found significant heterogeneity in the relative abundance of pathogens prior to infection for other bacteria. These findings demonstrate the utility of StrainSifter in strain matching between isolate and metagenomic sequencing data, and provide a more precise and comprehensive investigation of the intestine as a potential reservoir of diverse pathogens capable of causing bloodstream infections in HCT recipients. These findings may have implications for the tracking and prevention of BSI previously classified as central-line associated bloodstream infections.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.